Research
The Van Deventer Laboratory is broadly interested in how to engineer more “druglike” proteins. We make use of tools from protein engineering, synthetic biology, and chemical biology.
One major interest of the lab is in engineering protein-small molecule “hybrid” structures to disrupt the functions of biological targets poorly served by existing classes of therapeutics. We reason that integrating small molecule and protein functionality will enable us to simultaneously leverage the best properties of each of these classes of molecules. Toward this end, we are learning how to construct, evaluate, and screen hybrid structures on the yeast surface using the combination of yeast display and noncanonical amino acids. Our initial efforts in this area are focused on how best to present chemical functionality within antibodies and other proteins, and on the discovery of metalloproteinase inhibitors that can discriminate between closely related enzymes. A portion of these efforts focus on generating tools for understanding the biology of the tumor microenvironment.
We are also extremely interested in enhancing the process of genetically encoding noncanonical amino acids in proteins. In particular, in yeast, the tools for adding noncanonical amino acids to the genetic code remain underdeveloped. We have exploited reporter systems for quantifying noncanonical amino acid incorporation efficiency and fidelity. In addition, we expect these tools to support efforts to engineer the protein translation apparatus—and the yeast cells housing the apparatus—to push the capabilities of these methodologies in yeast to new heights. Engineering cells to better accommodate alternative genetic codes will enhance the “chemical versatility” that we can genetically encode, which has implications for our own hybrid engineering work and for broader topics ranging from basic biology studies to the engineering of biomaterials and therapeutic proteins.
Finally, we are exploring other strategies for making proteins more “druglike.” Our first investigations in this area are focused on intracellular delivery of proteins for the purpose of interfering with intracellular signaling. Enabling proteins (or hybrids) to readily access the cytosol could result in a number of new opportunities for disrupting biological targets that have traditionally been considered “undruggable.”
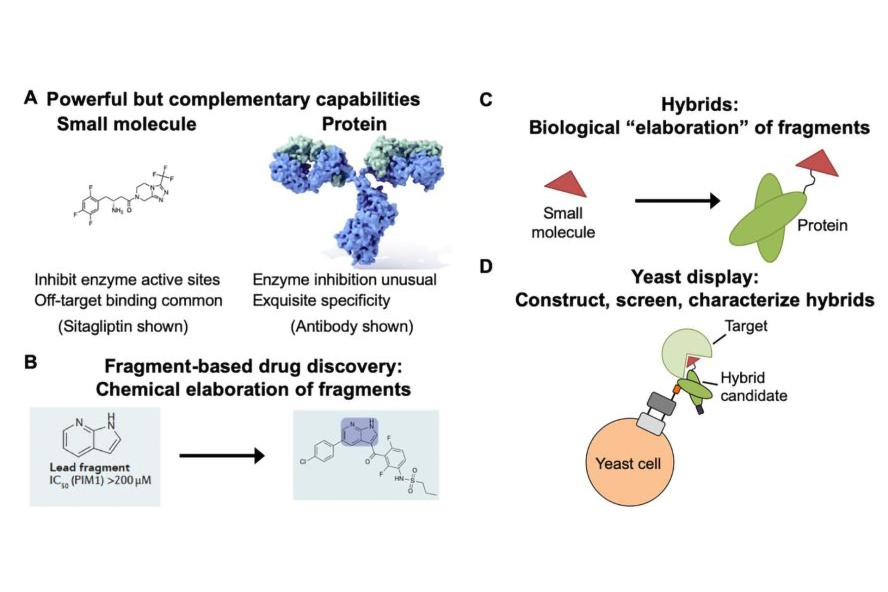
Figure 1: Hybrids as an approach to make proteins more “druglike.” A) Small molecules and proteins exhibit complementary properties as therapeutics. B) Chemical elaboration during conventional fragment-based drug discovery. C) Biological “elaboration” of small molecules with antibodies or other proteins. D) Preparing hybrids on the yeast surface as a strategy for high throughput screening and characterization. Graphics in (B) from Erlanson, D. A. et al. Nat Rev Drug Discov 2016, 15 (9), 605-19. Antibody image in (C) is from here.
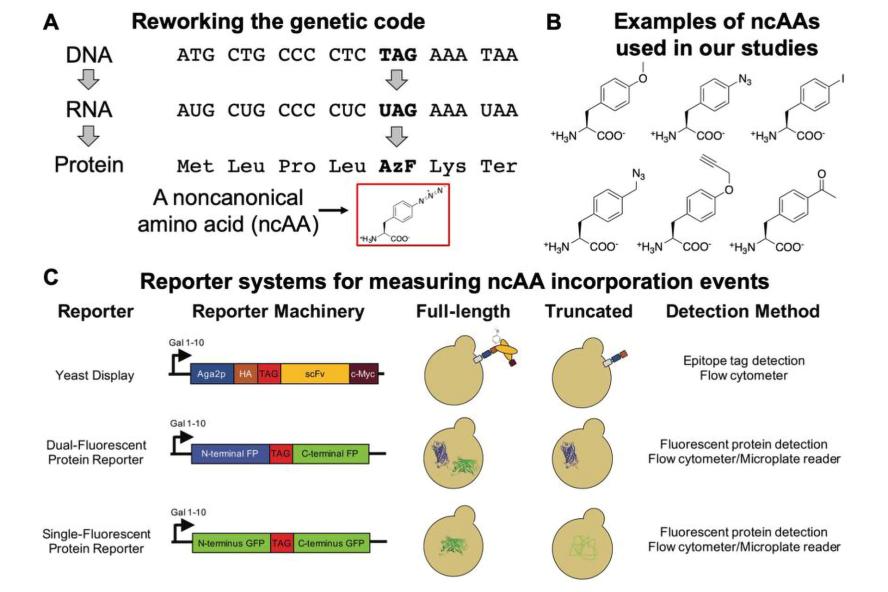
Figure 2: Changing the genetic code to encode noncanonical amino acids (ncAAs). A) Genetic code expansion to add a 21st amino acid to the genetic code. In this case, the “TAG” stop codon is being repurposed as a cognate codon. This requires the addition of engineered protein translation machinery to a cell (an additional aminoacyl-tRNA synthetase/tRNA pair, also called orthogonal translation system). B) Selected structures of noncanonical amino acids used in our studies. C) Reporter systems to quantify ncAA incorporation efficiency and fidelity in yeast. Measurements and analysis methods for evaluating these events are critical to understanding and enhancing the use of alternative genetic codes. Image is taken from here.