Development of novel optical biomarkers for the non-invasive detection of early cancer
Cancer is a leading cause of death worldwide. Elucidating the causes and underlying mechanisms directing cancer development is vital for designing effective detection and targeted treatment strategies. Our lab's research interests focus on employing high-resolution, depth-resolved imaging that relies entirely on endogenous contrast to non-invasively investigate cancer progression and develop novel quantitative, diagnostic biomarkers. The use of endogenous (i.e. natural) optical contrast obviates the need for exogenous agents that may be toxic or introduce artifacts and allow for a more straightforward path to human clinical applications.
Most cancers, including skin, breast, cervical, prostate, colon, ovarian and cervical cancer, develop in the epithelium, i.e. the surface/lining of the organ. While in the case of the skin and the oral cavity, these tissues are readily accessible, for other organs, such as the colon, the esophagus and the cervix, the epithelium is visualized using optical probes, such as endoscopes and colposcopes. Therefore, we already have means to access these tissues, even though they may not be very close to the surface of our bodies.
I. Engineered Epithelial Tissue Equivalents serving as a model of precancerous epithelial changes.
Collaborator:
Karl Münger, PhD, Harvard Medical School, Department of Pathology
Epithelia constitute one of the four basic animal tissue types. They cover or line organ surfaces and cavities and serve multiple functions, including protection, secretion and absorption. Non-invasive approaches to assess epithelial cancerous transformation could impact greatly clinical diagnostics as the majority of cancers are of epithelial origin. We have employed Engineered Epithelial Tissue Equivalents, constructed by either primary human foreskin keratinocytes (i.e. normal human epithelial cells) or immortalized human foreskin keratinocytes expressing the full-length genome of human papilloma virus 16 (HPV16), as a biological model to study HPV induced epithelial precancer. These are tissues that we can grow in the lab and they look very similar to normal human tissues. In fact, these are the types of tissues that are already commercially available for transplantation in patients with severe skin injuries, as a result of burns or ulcers, for example.
We studied those tissues non-invasively by utilizing different modes of multiphoton microscopic imaging, like Two-Photon Excited Fluorescence (TPEF) and Second Harmonic Generation (SHG). (Levitt JM et al, 2011, also see the following review articles for a more general perspective: Georgakoudi and Quinn, Annu Rev Biomed Eng 2012; Georgakoudi et al. Tissue Eng Part B Rev. 2008). Below are examples, of what the normal and pre-cancerous tissue equivalents look like using traditional images acquired by fixing, cutting and staining the tissue and the images we acquire without interfering with the tissue in any way.
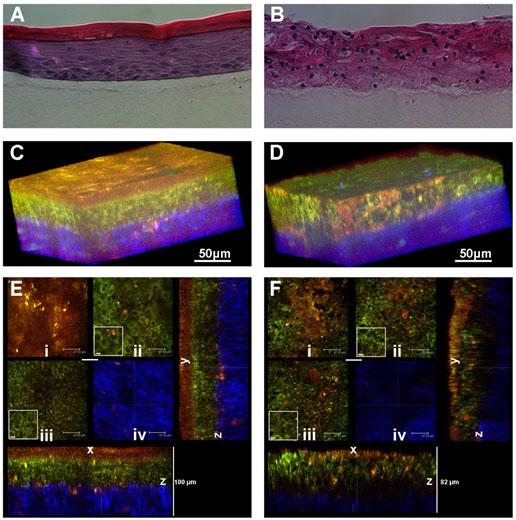
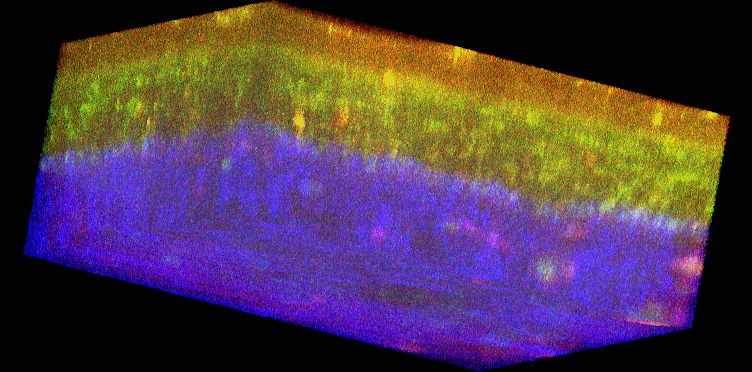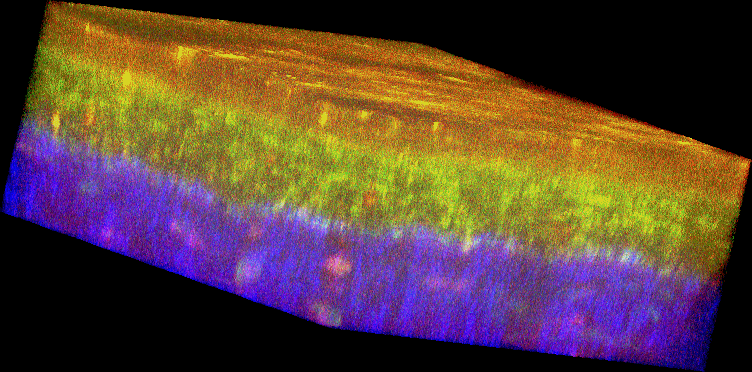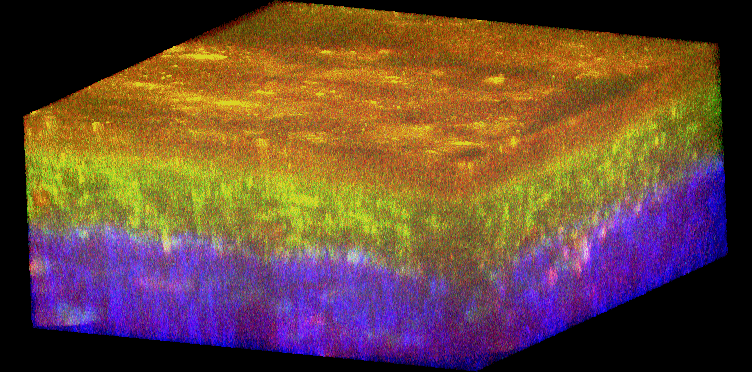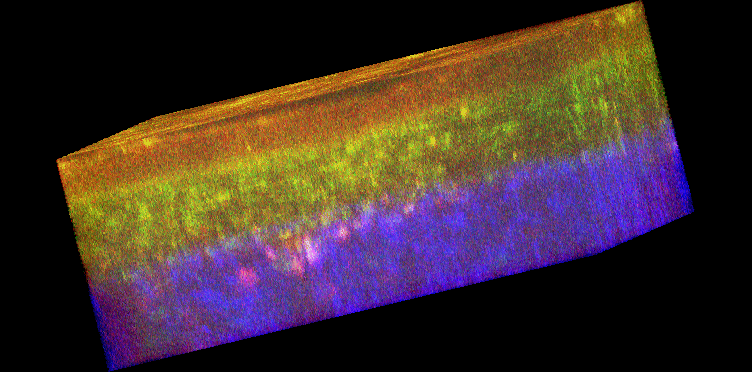
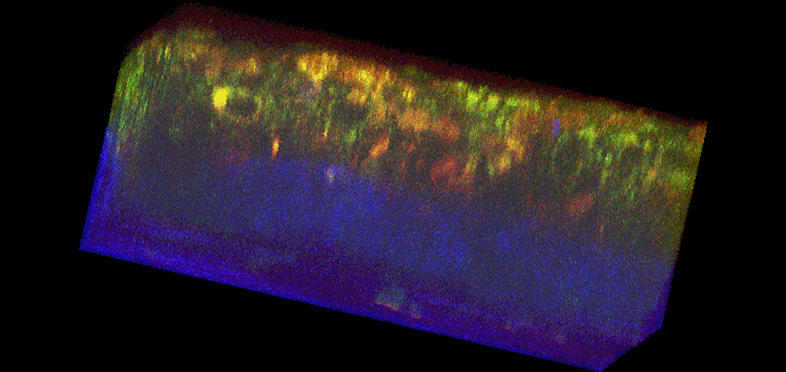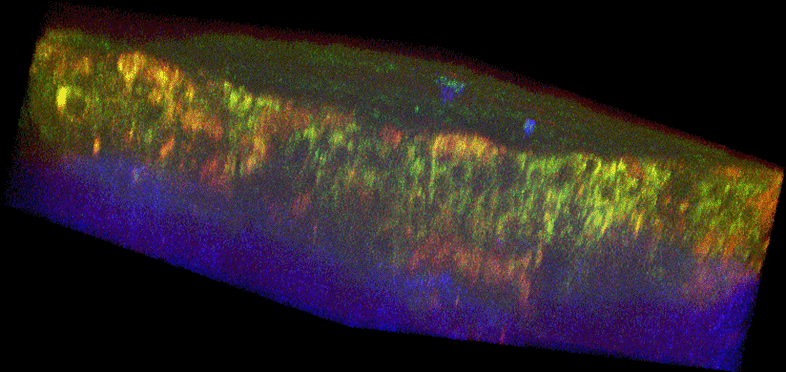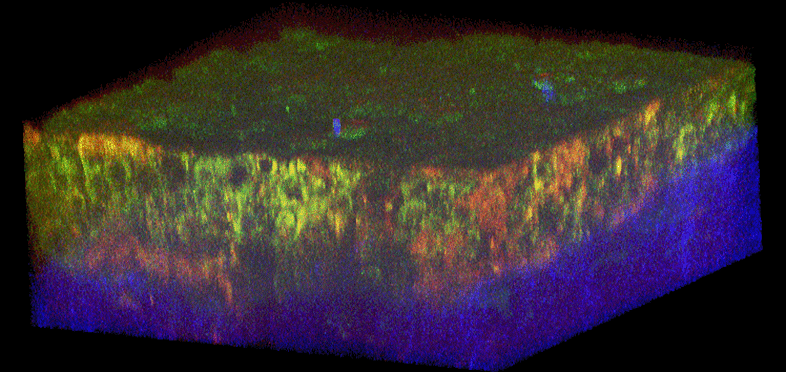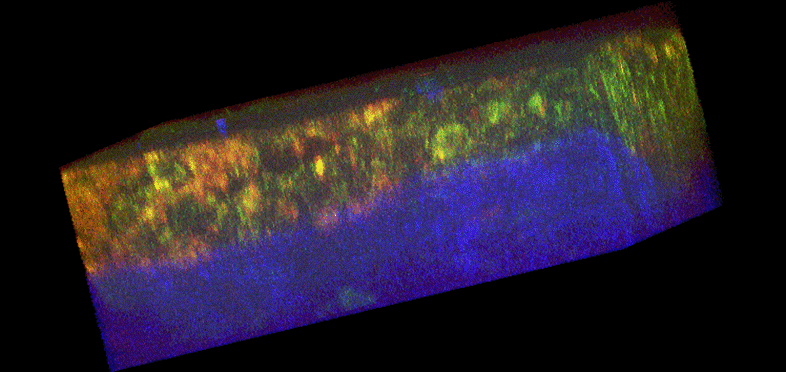
Figure 1. Histology and fluorescence based images of Engineered Epithelial Tissue Equivalents.
Normal (left column) and HPV- immortalized tissues (right column). Histological hematoxylin and eosin staining (a–b). False-color, depth-resolved TPEF and SHG images (c–f) from tissue excited at 755 nm and 800 nm. Green is fluorescence in the 455 nm channel (from NADH and keratin), while red is signal from the 525 nm channel (from FAD and keratin), acquired at 755 nm excitation. Blue represents 400 nm SHG detected at 800 nm excitation. Three-dimensional reconstructions were created from the fluorescence images (c–d). Corresponding depth images are shown in (e–f) with increasing depth from i to iv with zoomed insets to highlight the cell sizes. The scale bar represents 47.5 µm in the images and 10 µm in the zoomed insets. The transverse views of the normal and HPV-immortalized tissues (zx & zy) are shown with scale bars of 100 µm and 82 µm, respectively.
II. Quantitative characterization of tissue biochemistry and organization.
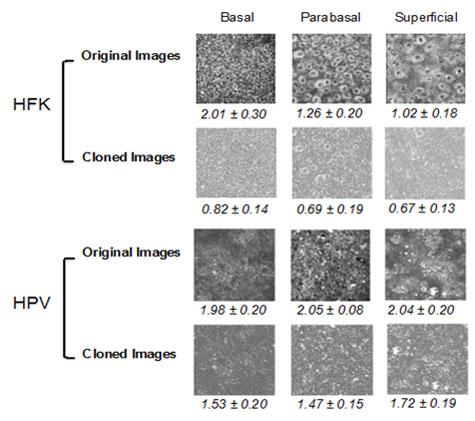
A great need exists for image assessment tools that will efficiently extract features and patterns from biomedical images in a quantitative and automated manner and will decrease or eliminate human subjectivity from the interpretation process. Ideally, such imaging approaches can be implemented in near real-time and provide quantitative biomarker outcomes to the physician at the time of examination. This would enable immediate treatment and would eliminate the stressful time that a patient often needs to wait for a diagnosis. Our lab is interested in developing such processing tools to analyze biomedical images. One area of focus is the fractal nature of subcellular inhomogeneities and their variation with disease state. Images of living cells and tissues obtained via fluorescence microscopy, due to their submicron scale resolution, provide a direct and noninvasive way to analyze subcellular morphology. We have developed an automated, Power Spectral Density (PSD) based methodology that overcomes limitations and artifacts introduced by the presence of cellular borders for quantitatively characterizing the subcellular organization of cellular autofluorescence (Levitt et al, 2007; Xylas J. et al, 2012). The technique is based on isolating cell features via thresholding and then applying a digital object cloning (DOC) method in order to produce images that contain only intracellular signal information. The figure below demonstrates the application of the aforementioned algorithmic process to normal (HFK) and diseased (HPV) tissue images. These images represent spatial variations in NADH fluorescence intensity throughout the cells, which in turn emanate primarily from mitochondria. Therefore the numeric value of the parameter included in these figures represents distinct levels of mitochondrial organization, which are significantly different between the normal and pre-cancerous tissues we examine. The images are acquired from live tissues without any need to process or stain. This type of analysis reveals a novel set of biomarkers than the ones that pathologists typically consider and could enhance significantly diagnostic capabilities.
Figure 2. Representative original and DOC-corrected TPEF images of superficial, para-basal, and basal layers of epithelial tissues made with healthy human foreskin keratinocytes (HFK) and HPV-transfected keratinocytes with the average values for 5 different fields for each group displayed under each image with standard deviations.
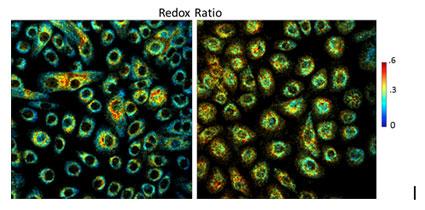
Another very intriguing facet of TPEF tissue images that our lab focuses on, is their capacity as carriers of biochemical information. Cells contain molecules that autofluoresce naturally when excited under appropriate conditions. Some of these molecules, such as NADH (nicotinamide adenine dinucleotide) and FAD (flavin adenine dinucleotide), are also enzymatic cofactors participating in the cells' key metabolic and biosynthetic pathways. This enables the utilization of fluorescence microscopy for biological imaging to investigate cellular metabolism. Because FAD and NADH are the only forms of electron carriers that exhibit endogenous fuorescence, a ratio of FAD/(NADH+FAD) fuorescence can be used to estimate the metabolic profile of live cells or tissues and provide insight into their metabolic state. Cells dynamically adjust their metabolism to satisfy their energetic requirements while they undergo normal (ie differentiation) or pathological (ie cancerous transformation) processes. Therefore, the redox ratio can be ultimately used to non-invasively translate fluorescence intensity to cellular function and allow over time, live understanding of cellular activity. The following figure illustrates the changes in redox we see upon overexpression of the oncoprotein E7 in normal human foreskin keratinocytes.
Figure 3. Cancerous transformation affecting the redox ratio. Redox ratio color coded images of healthy Human Foreskin Keratinocytes (HFK), left, and keratinocytes transfected with the HPV16 E7 oncoprotein (E7), right. Colorbar scale corresponds to redox ratio values.
III. Steps towards translation to the clinic.
Collaborators:
Hong-Thao Thieu, MD, Obstetrics and Gynecology, Tufts Medical Center
Michael House, MD, Obstetrics and Gynecology, Tufts Medical Center
Maria Garcia-Moliner, MD, Pathology and Laboratory Medicine, Tufts Medical Center
We have found that non-linear microscopy is a technique that can be used to identify metabolic and organizational differences between normal and pre-cancerous engineered epithelial tissues. The next step involves assessment and further development of these tools to characterize human tissues. Therefore, we have started a strong collaboration with clinicians at the Tufts Medical Center that gives us access to freshly excised normal and pre-cancerous human cervical tissues. Cervical cancer is the second most common cancer affecting women worldwide, leading to hundreds of thousands of deaths, especially in the developing world. This is cancer that is almost always the result of HPV infection. Cancer morbidity and mortality can be prevented if lesions are detected and treated in their precancerous stage. We expect that the tools we are developing will assist in improving detection at this early stage.