Optical Monitoring of Cell-Matrix Interactions in Engineered Tissues
Biochemical, morphological and organizational changes Non-destructive characterization of collagen Monitoring of circulating cells in vivo or within microfluidic devices Tissue engineering requires the development and optimization of complex cell culture protocols in order to create functional three-dimensional tissues from biomaterial scaffolds seeded with stem cells.
Traditionally, the biochemical and structural characteristics of engineered tissues have been assessed through destructive lab techniques such as histology and quantitative PCR. Our primary goal is to develop non-destructive, label-free methods to assess the in vitro development and in vivo integration of engineered tissue implants.
The two main tissue aspects that we aim to characterize optically include cellular metabolism and collagen organization. Cell metabolism plays a central role in the regulation of normal development, function and death of a cell. Interestingly, there are two co-enzymes involved in key metabolic pathways, NADH and FAD, that naturally absorb light in the visible range of the spectrum and re-emit it at slightly longer wavelengths. These signals can be detected and quantified to assess the metabolic state of the cells. This information in turn can be used to assess normal or diseased cell development.
Collagen is the most prevalent protein in our bodies and serves multiple functions, such as providing support and a means to contain the different tissue components. Especially during the last few years it has become clear that the interactions between cells with their surrounding matrix, which often is rich in collagen, play a key role in normal and diseased tissue development. Collagen has also very unique optical properties, that enable us to image it, especially when in its fibrilar form, without the need to stain it. Therefore, through the use of appropriate methods and image analysis algorithms we can acquire very detailed information regarding collagen structure and organization and the changes that occur as a tissue develops normally or when a disease evolves.
High-resolution non-linear microscopy techniques, such as multi-photon excited fluorescence (MPEF) and second harmonic generation (SHG), offer the ability to probe a variety of endogenous sources of contrast. In collaboration, with Professors David Kaplan and Kyongbum Lee, we have been developing quantitative assessments of stem cell differentiation based on a redox ratio of NADH and FAD fluorescence. Through live cell imaging of these endogenous fluorophores, dynamic changes in the metabolic state of stem cells are observed during the early stages of differentiation. Additionally, MPEF and SHG imaging can be used to define changes in the microstructural organization of extracellular matrix proteins, such as collagen, keratin, and elastin. In collaboration with Dr. Emmanuel Beaurepaire at École Polytechnique, we have also developed techniques to non-invasively monitor lipid droplet formation in developing engineered adipose tissues using third harmonic generation microscopy.
The intrinsic optical sectioning capability and the deeper penetration of near-infrared light make the nonlinear optical techniques we employ in the lab particularly amenable to the assessment of 3D tissues. Recent work on this project has focused on quantifying the spatiotemporal dynamics of epithelial and stem cell interactions with collagen and silk-based scaffolds. Through the development of custom-written image analysis software in Matlab, cells can be automatically segmented from the surrounding scaffold. The automatic quantification of the metabolic status, morphology, and location of every cell within a 3D tissue volume can provide a rapid and comprehensive assessment of tissue development.
Related select publications
Hayden RS, Fortin J-P, Harwood B, Subramanian B, Quinn KP, Georgakoudi I, Kopin AS, Kaplan DL. Cell-tethered ligands modulate bone remodeling by osteoblasts and osteoclasts. Advanced Functional Materials. In Press.
Chang T, Zimmerley M, Quinn KP, Lamarre-Jounne I, Kaplan DL, Beaurepaire E, Georgakoudi I. Non-invasive monitoring of cell metabolism and lipid production in 3D engineered human adipose tissues using label-free multiphoton microscopy. Biomaterials. In Press.
Ward A, Quinn KP, Bellas E, Georgakoudi I, Kaplan DL (2013). Noninvasive metabolic imaging of engineered 3D human adipose tissue in a perfusion bioreactor. PLoS One 8(2): e55696.
Quinn KP, Bellas E, Fourligas N, Kaplan DL, Georgakoudi I (2012). Characterization of metabolic changes associated with the functional development of 3D engineered tissues by non-invasive, dynamic measurement of individual cell redox ratios. Biomaterials 33(21): 5341-8.
Georgakoudi I, Quinn KP (2012). Optical imaging using endogenous contrast to assess metabolic state. Annual Review of Biomedical Engineering 14: 351–67.
Hronik-Tupaj M, Rice WL, Cronin-Golomb M, Kaplan DL, Georgakoudi I (2011). Osteoblastic differentiation and stress response of human mesenchymal stem cells exposed to alternating current electric fields. Biomed Eng Online 10: 9.
Rice WL, Kaplan DL, Georgakoudi I (2010). Two-photon microscopy for non-invasive, quantitative monitoring of stem cell differentiation. PLoS One 5(4): e10075.
Collaborators
Jonathan Garlick, Tufts School of Dental Medicine, Cell, Molecular & Developmental Biology
David Kaplan, Tufts University, Department of Biomedical Engineering
Kyongbum Lee, Tufts University, Department of Chemical and Biological Engineering
Karl Munger, Harvard Medical School, Department of Pathology
Emmanuel Beaurepaire, Laboratory for Optics and Biosciences, École Polytechnique.
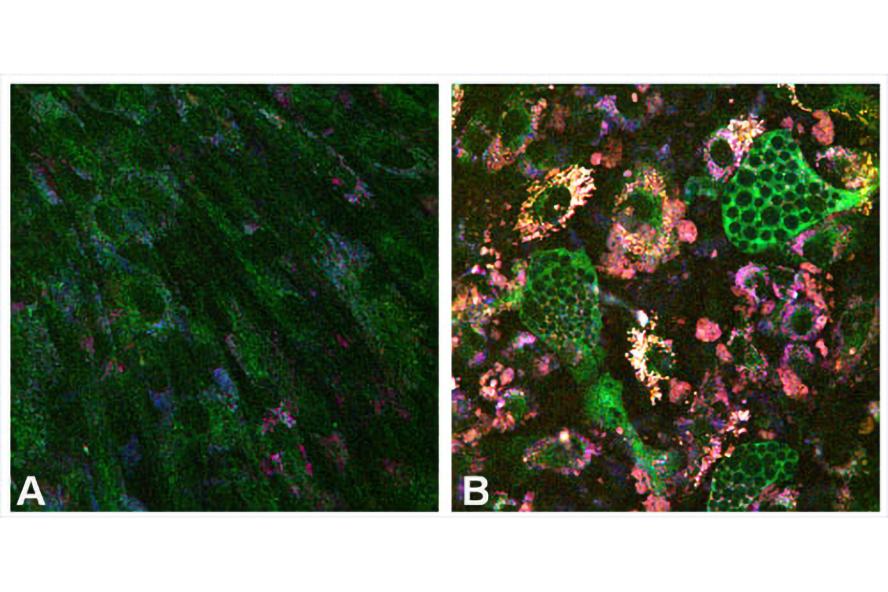
Figure 1. NADH (green), FAD (blue), and lipofuscin (red) fluorescence differ among (A) propagating mesenchymal stem cells (MSCs) and (B) MSCs that underwent adipogenic differentiation.
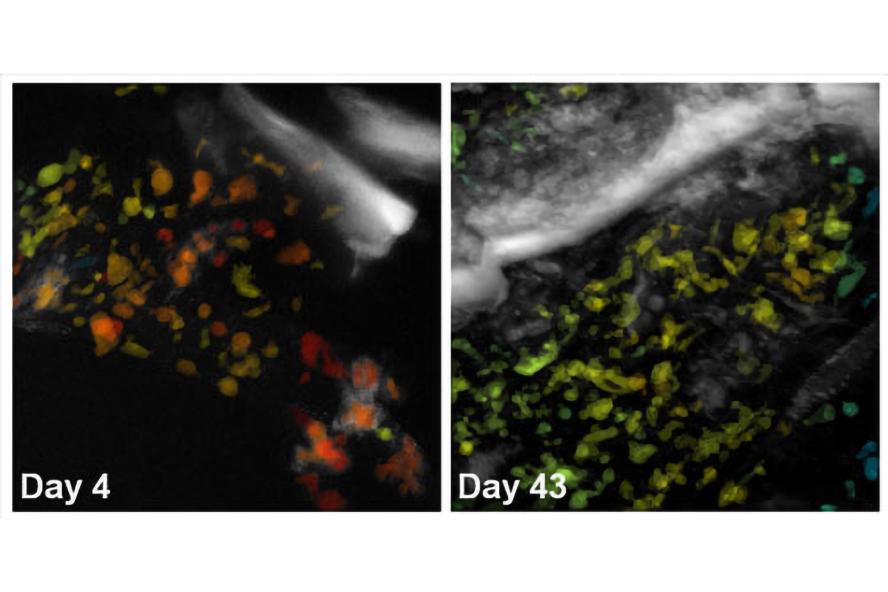
Figure 2. Cells undergoing adipogenic differentiation within a 3D silk scaffold have been automatically segmented and color-coded for their average optical redox ratio. Z-projections of two image volumes demonstrate changes in cell redox state over time.
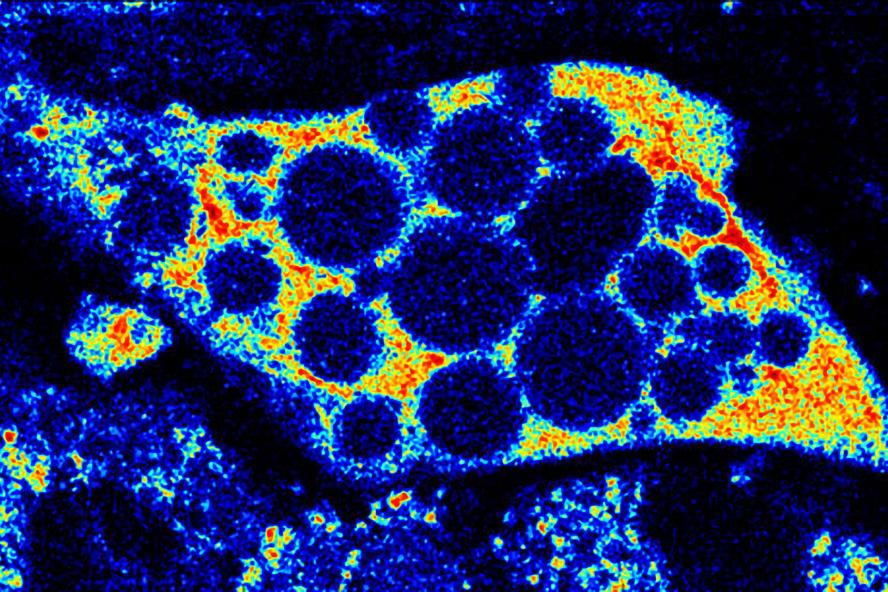
Figure 3. 1st Place VRTA photography competition. NADH fluorescence can be used to assess the mitochondrial network of a maturing adipocyte. By assessing the spatial organization and relative changes in NADH intensity, these two-photon images are used to non-destructively assess cell differentiation status and metabolic activity within in vitro cell cultures.

Movie 1. 3D projection of silk scaffold (grayscale) embedded with hMSCs and treated with adipogenic media for nine weeks. Cell redox ratios calculated based on recorded endogenous 2PEF images are represented by the jet color map. Lipid droplets detected in corresponding THG images are shown in magenta. Adapted from Chang et al., in press.