Engineering enzymes in high throughput
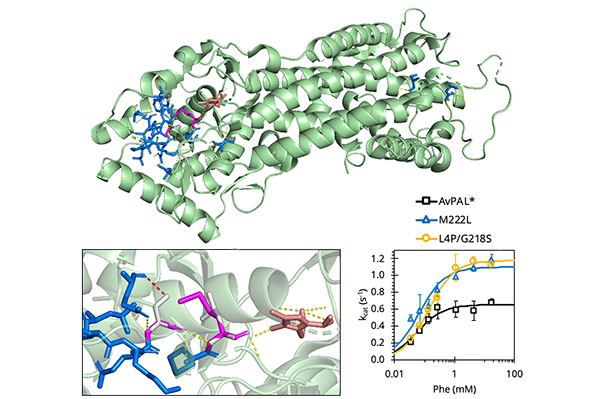
Phenylalanine ammonia-lyase (PAL) is a class of enzymes with an array of potential biocatalytic applications in industry and in medicine – including as a therapeutic enzyme in cancer treatment or in the treatment of phenylketonuria (PKU), an inborn error of metabolism that can lead to seizures, behavioral problems, intellectual disability, and mental disorders.
Researchers have previously been able to improve PAL stability or substrate specificity by using laboratory procedures to drive metagenesis, modifying the enzyme’s genetic information with a mutation. That process has been site-specific, though, and combinatorial techniques have not been widely explored.
In the Tufts Department of Chemical and Biological Engineering, the Synthetic Biology and Systems Bioengineering Lab developed a directed evolution technique to engineer PAL enzymes, opening the way for the exploration of a more comprehensive mutational landscape. In research reported in Chemical Communications, the team coupled E. coli growth with PAL activity for high-throughput enrichment.
Working with the clinically-relevant PAL used in the formation of pegvaliase, the first enzyme substitution therapy for patients with PKU approved by the U.S. Food and Drug Administration, the researchers found previously unidentified mutations, which “increase turnover frequency almost twofold after only a single round of engineering.”
PhD candidate Zachary Mays and postdoctoral scholar Dr. Karishma Mohan were co-first authors on the paper. They worked with Dr. Vikas Trivedi, PhD candidate Todd Chappell, and corresponding author Assistant Professor Nikhil Nair.
“There is so much we cannot yet predict when engineering a complex macromolecule like an enzyme,” says Mays. “It’s exciting to screen a combinatorial library of mutations, most of which would be overlooked when using a targeted approach and find unexpected candidates that improve activity.”
Department:
Chemical and Biological Engineering